Is SARS-CoV-2 a bacteriophage?
Article
Carlo Brogna 1&*, Barbara Brogna 2, Domenico Rocco Bisaccia 1, Francesco Lauritano 1, Marino Giuliano 3, Luigi Montano 4, Simone Cristoni 5, Marina Prisco 6, and Marina Piscopo 6&*.
1. Department of research, Craniomed group facility Srl., Montemiletto (Av), Italy; dir.brogna@craniomed.it (C.B.); roccobisa@gmail.com (R.D.B.); francescodrlauritano@gmail.com (F.L.)
2. Department of Radiology, Moscati Hospital, Contrada Amoretta, 83100 Avellino, Italy; brognabarbara1@gmail.com (B.B.)
3. Marsanconsulting Srl. Public Health Company; Via dei Fiorentini, 80133 – Napoli; Italy; marino@marsanconsulting.it (M.G.)
4. Andrology Unit and Service of LifeStyle Medicine in Uro-Andrology, Local Health Authority (ASL) Salerno; luigimontano@gmail.com (L.M.)
5. ISB – Ion Source & Biotechnologies Srl- Bresso Milano; simone.cristoni@gmail.com (S.C.)
6. Department of Biology, University of Naples Federico II, 80126 Napoli, Italy; marina.prisco@unina.it (M.P.); marina.piscopo@unina.it (M.P.)
* Correspondence and equal contributor: marina.piscopo@unina.it; dir.brogna@craniomed.it
Abstract: SARS-CoV-2 has become one of the most studied viruses of the last century. It was assumed that the only possible host for these types of viruses was mammalian eukaryotic cells. Our recent studies show that microorganisms in the human gastrointestinal tract affect the severity of COVID-19 and demonstrate for the first time that the virus replicates in gut bacteria. To complete this investigation, cultures of bacteria from the human microbiome and SARS-CoV-2 were analyzed by electron and fluorescence microscopy, in this work. The images presented in this paper, in association with the Nitrogen (15N) isotope-labeled culture medium experiment, clearly show that SARS-CoV-2 infects bacteria in the gut microbiota, indicating that SARS-CoV-2 acts as a bacteriophage. Our results add new knowledge to the understanding of the mechanisms of SARS-CoV-2 infection and fill gaps in the study of the interactions between SARS-CoV-2 and non-mammalian cells. These conclusions could suggest specific new pharmacological solutions to support the vaccination campaign.
Keywords: SARS-CoV-2; human microbiota; Sabin vaccine; electron microscope; mucosal immunity; bacteriophage
1. Introduction
Coronavirus disease 2019 (COVID-19) is having a devastating effect on the world’s
population, as it has caused more than 5.4 million deaths worldwide. For this reason, this
disease is recognized as the most significant global health crisis since the 1918 influenza
pandemic. Since the World Health Organization (WHO) declared a global pandemic on March
11, 2020, many countries continue to experience multiple waves of outbreaks of this viral
disease.
Currently, a new variant has already arrived from South Africa: “Omicron.” The data
officialized by the WHO as of November 30, 2021, are as follows: 261,435,768 confirmed
cases; 5,207,634 deaths; 7,772,799,316 doses of vaccine administered [1]. The vaccines
administered are almost half of the “classic” vaccines based on inactivated viruses
(CoronaVac produced by SinoVac), and half of the latest generation of vaccines based on
adenoviral vectors (AZD1222 by AstraZeneca, Ad26.COV2.S by Janssen) or mRNAs (
mRNA-1273 by Moderna, BNT162b2 by Pfizer – BioNTech) [1]. In terms of vaccinated
individuals, about 4 billion people worldwide have received a dose of vaccine (about 47.3%
of the world population): 40% of them have completed a whole cycle of two vaccinations, and
a small group (0.5%) has finished the third booster (as in Israel). Many studies since the
beginning of the pandemic have focused on the interaction between the ACE2 receptor and the
virus spike glycoprotein [2], how the enzyme furin works, and how it plays a critical role
between the eukaryotic host cell and the viral particle [3]. These observations are derived from
past knowledge of SARS [4] and MERS [4]. However, has it been tested whether these RNA
viruses, including coronaviruses, can also replicate in cells other than the mammalian
eukaryotic cell? In addition, has it been investigated whether they also replicate in yeast or
bacteria? [5]. Our previous work showed that people with COVID- 19 had toxin-like peptides,
which were almost identical to the toxic components of animal venoms, such as conotoxins,
phospholipases, phosphodiesterases, zinc metal proteinases, and bradykinins, in both blood,
feces, and urine [6]. In a later study, we demonstrated spontaneous replication of SARS-CoV-
2 in bacterial cultures from patient feces for up to 30 days and beyond [7]. The European
Commission, i.e. the Joint Research Centre (JRC) had made these new findings official with
the following study “A new JRC study shows however, and for the very first time, that SARSCoV-
2 may replicate in the bacteria of our guts” [8]. For that matter, there have also been
other researchers who have studied the relationship between the gut microbiota and COVID19.
Wang et al., 2021, for example, found significant alterations in the gut and respiratory
microbiome in COVID-19 patients, finding that there was an increase in pathogenic bacteria
and a decrease in the beneficial ones [9]. In addition, to further understand microbial function
in the lower respiratory tract in COVID-19 infection, a research group of San Jose, CA,USA,
conducted a functional analysis of previously published metatranscriptome sequencing data of
bronchoalveolar lavage fluid from COVID-19 cases, patients with community-acquired
pneumonia, and healthy controls [10]. By investigating related metabolic pathways,
distinguishable functional signatures in COVID-19 respiratory tract microbiomes were
revealed, including decreased potential for lipid metabolism and glycan biosynthesis and
metabolism pathways, and enhanced potential for carbohydrate metabolism pathways. The
findings also suggested new connections to consider, possibly specific to the lower respiratory
tract microbiome, requiring further research on microbial function and host-microbiome
interactions during SARS-CoV-2 infection [10]. Another very interesting finding emerged
from a bioinformatics analysis that revealed that some members of commensal upper
respiratory tract (URT) bacteria express proteins that bind the SARS-CoV-2 spike
glycoprotein, for example the ACE2-like protein. Based on this analysis and available data
showing a decline in the population of these bacteria in the elderly, it has also been proposed
that some commensal bacteria of the upper respiratory tract prevent SARS-CoV-2 infectivity
and that a decrease of these bacteria contributes to infection severity [11]. The same authors,
in another study observed that N501Y mutations in the receptor-binding domain (RBD) of the
SARS-CoV2 spike glycoprotein may allow increased binding to the ACE2 receptor when
natural products (NPs) of upper respiratory tract bacteria are present [12]. In addition, there is
also another thing to consider. Bacteriophages are viruses that infect bacterial cells. These
viruses have long been considered neutral to animals and humans because specific receptors
for bacteriophages on eukaryotic cells are lacking. However, very recent studies have provided
clear evidence that bacteriophages can interact with eukaryotic cells, causing effects on the
functions of the immune system, respiratory system, central nervous system, gastrointestinal
system, urinary tract, and reproductive system [13].
Given these assumptions, in order to add new knowledge to the understanding of the mechanisms of SARS-CoV-2 infection and to fill the gaps in the study of the interactions between SARS-CoV-2 and non-mammalian cells, in the present work, bacterial and SARSCoV-2 culture preparations, obtained from the experiment published in our first work [7], were analyzed by electron and fluorescence microscopy. The aim of our study was to elucidate the interaction between SARS-CoV-2 and human gut microbiota. The data will help to suggest specific therapeutic approaches to support the current vaccines and improve the vaccination campaign.
2. Results
2.1 Replication and viral load increase of SARS-CoV-2 in bacterial cultures
The curves of replication and viral load increase of SARS-CoV-2 in bacterial cultures are
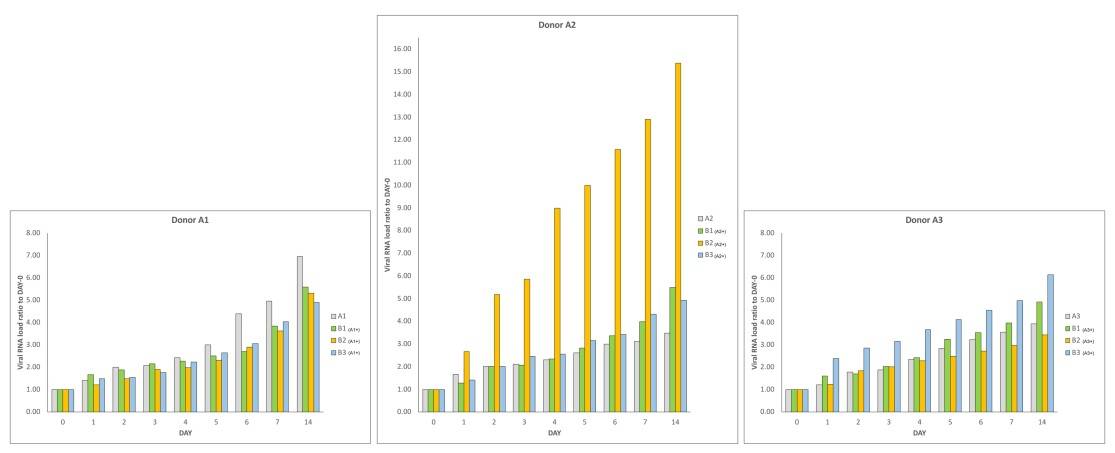
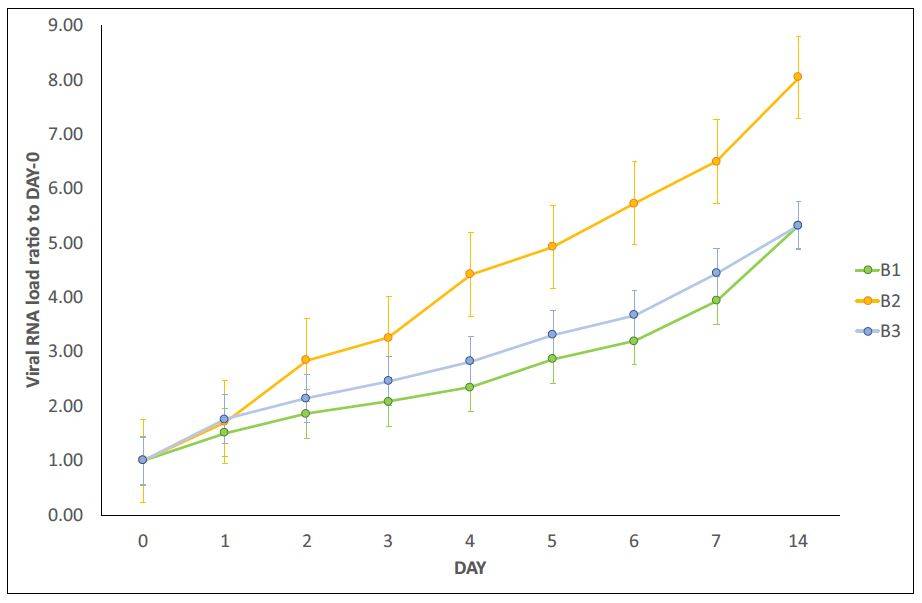
shown in Figure 1, panels A and B, with gentle permission of Dr. Petrillo [7]. In particular,
three couples of faecal samples from different “infected donors” (i.e. sources of A) and
“healthy recipients” (i.e. sources of B) have been recruited, and subject to the same
experimental procedure described in materials and methods. All combinations of “infected
donors” sources (A1, A2 and A3) and “healthy recipients” donors (B1, B2 and B3) were
subject to the same experimental procedure. Although with certain differences, the observed
trends were similar (Figure 1A), confirming the increase over time of SARS-CoV-2 RNA load
in samples of type A and in samples of type B(A+). Lastly, for each “recipient”, SARS-CoV-
2 RNA load was measured (Figure 1B). The results report virus replication in bacterial
cultures suggesting that this virus behaves as a bacteriophage.
2.2. Microscopic analysis of the interaction between SARS-CoV-2 and the human microbiome
To deepen our previous studies [6, 7], analyses using immunofluorescence, TEM, and
immunogold marking techniques were conducted. These analyses showed infection of
bacteria by SARS-CoV-2 virus after 30 days of bacterial culture, where mammalian
eukaryotic cells are excluded, in agreement with current literature that excludes their survival
beyond 48 hours [14]. Culture conditions have been described previously (see Materials and
Methods by Petrillo et. al. [7]).
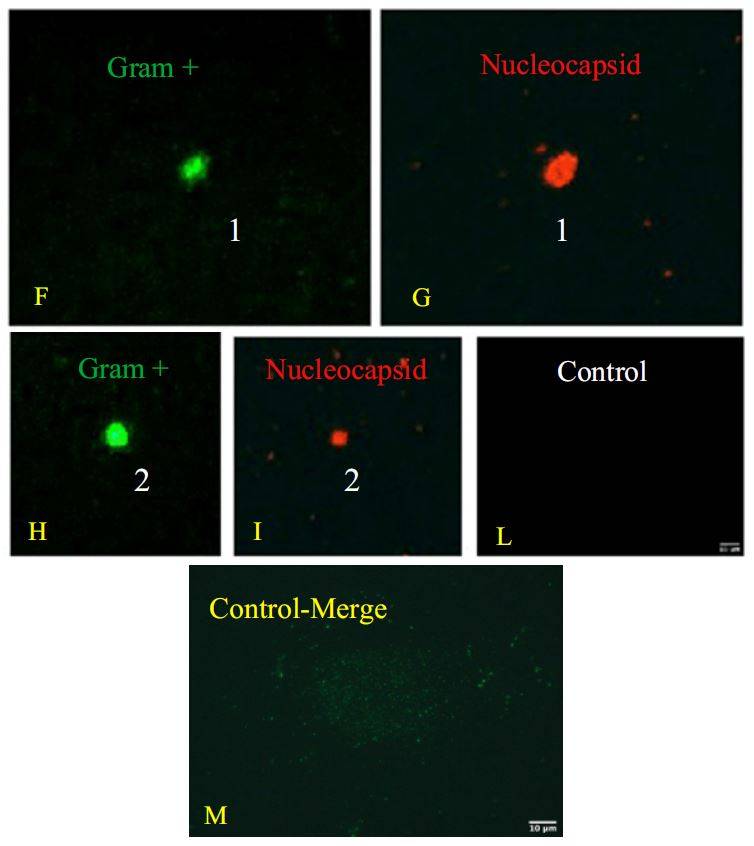
Double immunofluorescence was conducted using anti-Gram Positive Bacteria Marker
and anti-SARS-CoV-2 nucleocapsid protein and this experiment showed co-localization of
both signals on bacterial cells (Fig.1).In particular, the green signal corresponds to the Grampositive
bacteria that are shown in the images in panels C, F, H. The red signal indicates the
nucleocapsid protein of SARS-CoV-2. In panels D, G, and I, the same bacteria shown in the
previous panels are shown. Since these are labeled with this signal, it means that they are
infected with the virus. The confirmation is obtained from the overlay/merge fluorescence
shown in the panel E in which the gram-positive bacteria (signal with green light- in panel C,
F, and H) were attached to viruses like-particles of SARS-CoV-2 (signal with red light-, in
panel D, G, and I). In panels F and H it is possible to observe the computerized magnifications
of the bacteria indicated with numbers 1 and 2 that appear green because they are marked with
the anti Gram-positive antibody. Similarly, panels G are computerized magnification of the
bacteria attached by SARS-CoV-2 indicated with the numbers 1 and 2. The negative control
of the culture obtained without primary antibodies but only with secondaries antibodies is
shown in panel L. Panel M is the merge of the negative control of a healthy fecal bacterial
culture, in which the antibodies versus nucleocapsid protein didn’t give signal and Grampositive
bacteria are present.
An analysis performed by transmission electron microscopy, showed virus-like particles
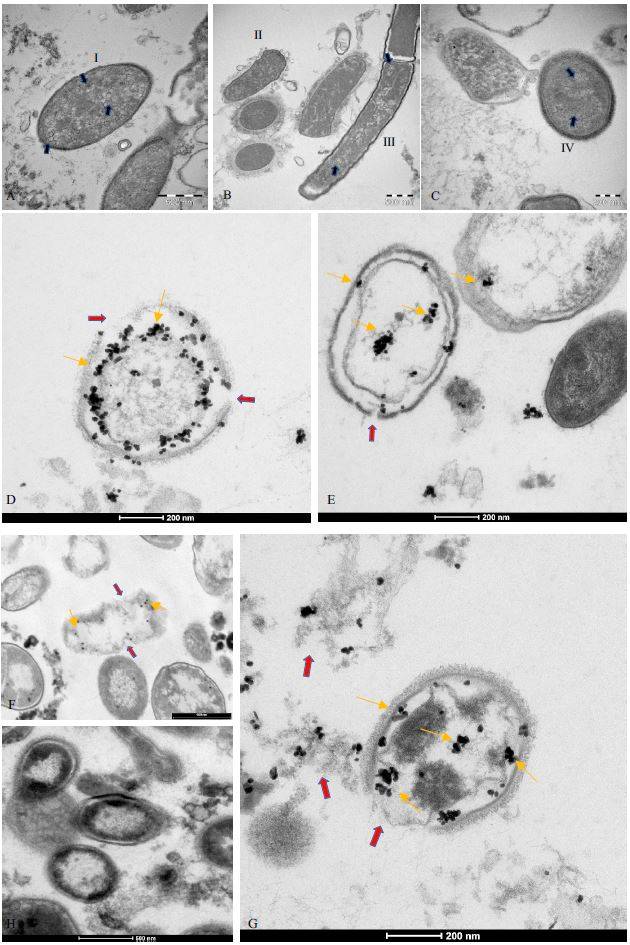
near and within the bacteria as can be seen in Fig.2 A-C. In addition, immunogold labeling
performed with anti-SARS-CoV-2 nucleocapsid protein antibody revealed the gold
nanoparticles in the bacteria (Fig. 2 D-G). Initial areas of cell wall lysis and fully lysed bacteria
are also observed (indicated in Fig. 2 with red arrows). Panel H represents the negative
immuno-gold control. No mammalian eukaryotic cells are present after 30 days of bacterial
cultures incubated under the conditions previously described in Petrillo et al. [7]. The presence
of gold nanoparticles in bacteria was also observed by performing immunogold preembedding
in another facility, using anti-SARS-CoV-2 nucleocapsid protein antibody. Only
one yeast was found in the culture after 30 days, and no viral particles were present (Fig.1
panel E-F of Supplementary Material). Visualization of gold nanoparticles in bacteria are clear
evidence that the virus is inside the bacteria.
Figure 1. Panel A-B: viral increase of SARS-CoV-2 in bacterial cultures. These images are reused and correspond to figure 3 of Petrillo et al. 2021 (doi: 10.12688/f1000research.52540.1.) obtained with gentle permission of Dr. Petrillo et al. 2021[7]. Results of experiments combining samples from three “infected donor” sources (A1, A2 and A3) and from three “healthy recipient” sources (B1, B2 and B3). A) The graphs report results of nine combinations. To normalize the measurements, all values at day 0 were used as the denominator (at day 0 all values = 1), i.e. for each sample, at day X, the ratio between Luminex Count At DayX/Luminex CountAt Day 0 was calculated. Each bar represents the SARS-CoV-2 RNA load ratio. B) Each line represents the average of the SARS-CoV-2 RNA load ratio of samples B1 (green line), B2 (yellow line), and B3 (azure line) infected each one with three different A donor sources. To normalize the measurements, all values at day 0 were normalised as described in panel A). Panels C-L show the immunofluorescence microscope images (Zeiss Axioplan 2, Axiocam 305 color, magnification 100X) on bacterial cultures (performed at 30 days) from the patients positive to SARS-CoV-2 with anti-gram-positive bacteria (green signal, C) and anti-SARS-CoV-2 nucleocapsid protein (red signal, D) antibodies. E: merge of C and D images, particles positive to both antibodies were indicated by white arrows. F-I: computerized magnification of particles numbered in C-E. Panel L: negative control by omitting primary antibodies. Panel M is the merge of the negative control of a healthy fecal bacterial culture, in which the anti-bodies versus nucleocapsid protein didn’t give signal and gram+ bacteria are present.
See text for more details.
Figure 2. TEM images of bacterial cultures from SARS-CoV-2 positive patients. A-C shows circular structures (blue arrows) inside bacteria cells (I, II, III, IV). D-G: immunogold labelling tecnique on the same samples with anti-nucleocapsid protein of SARS-CoV-2 antibody (gold arrows); the gold particles are inside the bacteria. Red
arrows show lysis of the bacteria membrane. Specifically, image G shows bacteria wall membrane lysate (red arrow) and the antibody binding to SARS-CoV-2 N protein in the cell (gold arrows). H: negative control.
See text for more details.
2.3 NGS sequencing data
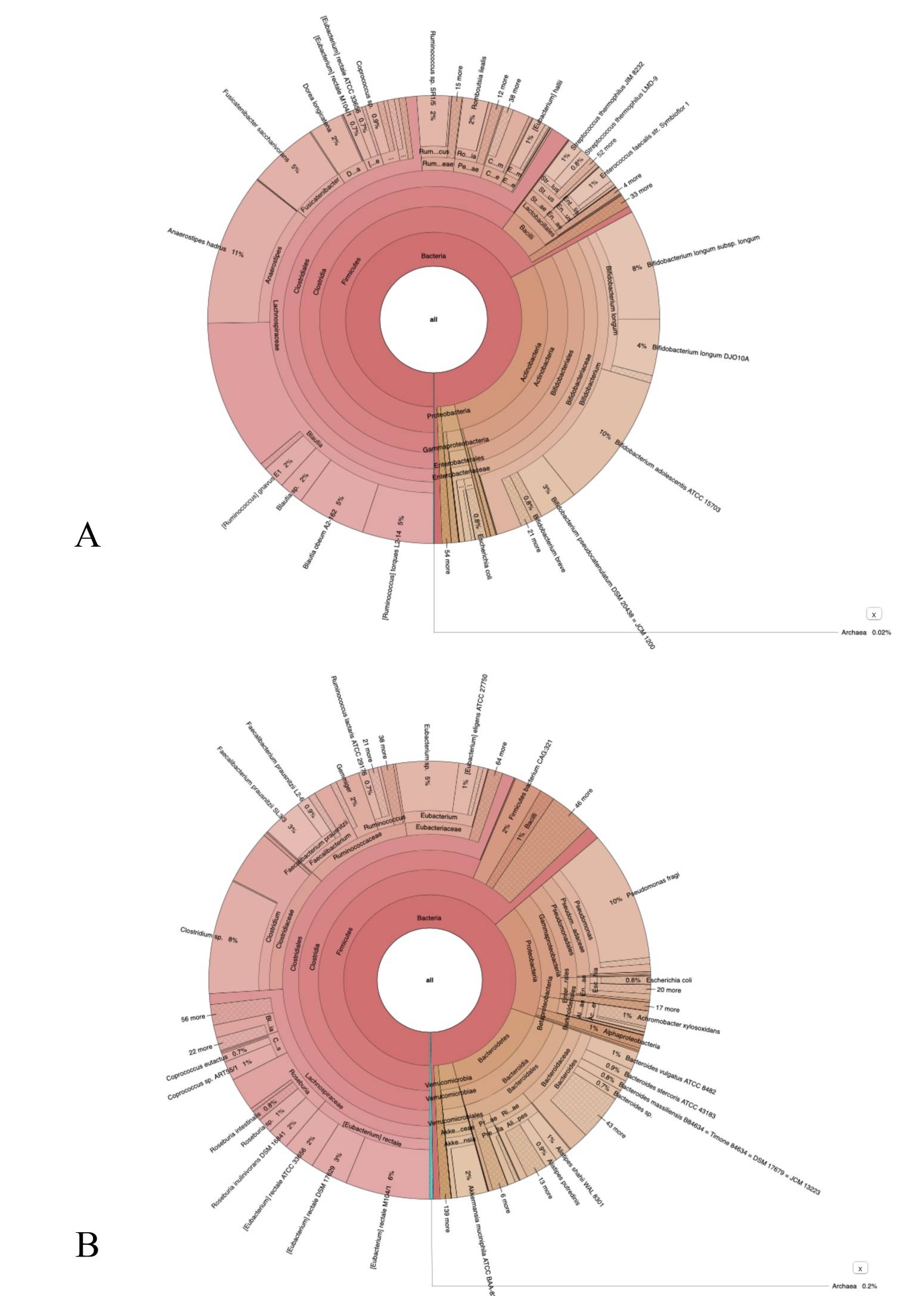
An interactive metagenomic visualization using the tool Krona [15] is shown in Figure 3 A-B. The multilayer pie chart represents the degrees of the bacterial world. The division is from inside to outside the circle. Colors towards red, have a low level of confidence while colors towards green have a high level of confidence. The initial and final samples, after 30 days, of the bacterial cultures are compared. The genera of bacteria such as Dorea, Fusicatenibacter, Klebsiella, Streptococcus decreased while other genera of bacteria such as Campylobacter, Prevotella, Staphylococcus, Bacteroides, and Cytobacter increased. In Table 2 (supplementary material), we provide a list of other bacteriophages present after 30 days of bacterial culture. These bacteriophages differ in size, shape, and most importantly are characterized by the presence of a tail, which is absent in the viral particles we present in TEM and immunogold images. Therefore, these data give insight into which genera of bacteria can undergo SARS-CoV-2 lysis and in particular make it possible to exclude other bacteriophages characterized by accessory microscopic structures that the virions shown do not possess.
2.4 Nitrogen isotope 15N assay in bacterial culture medium. Proteomic profiling analysis.
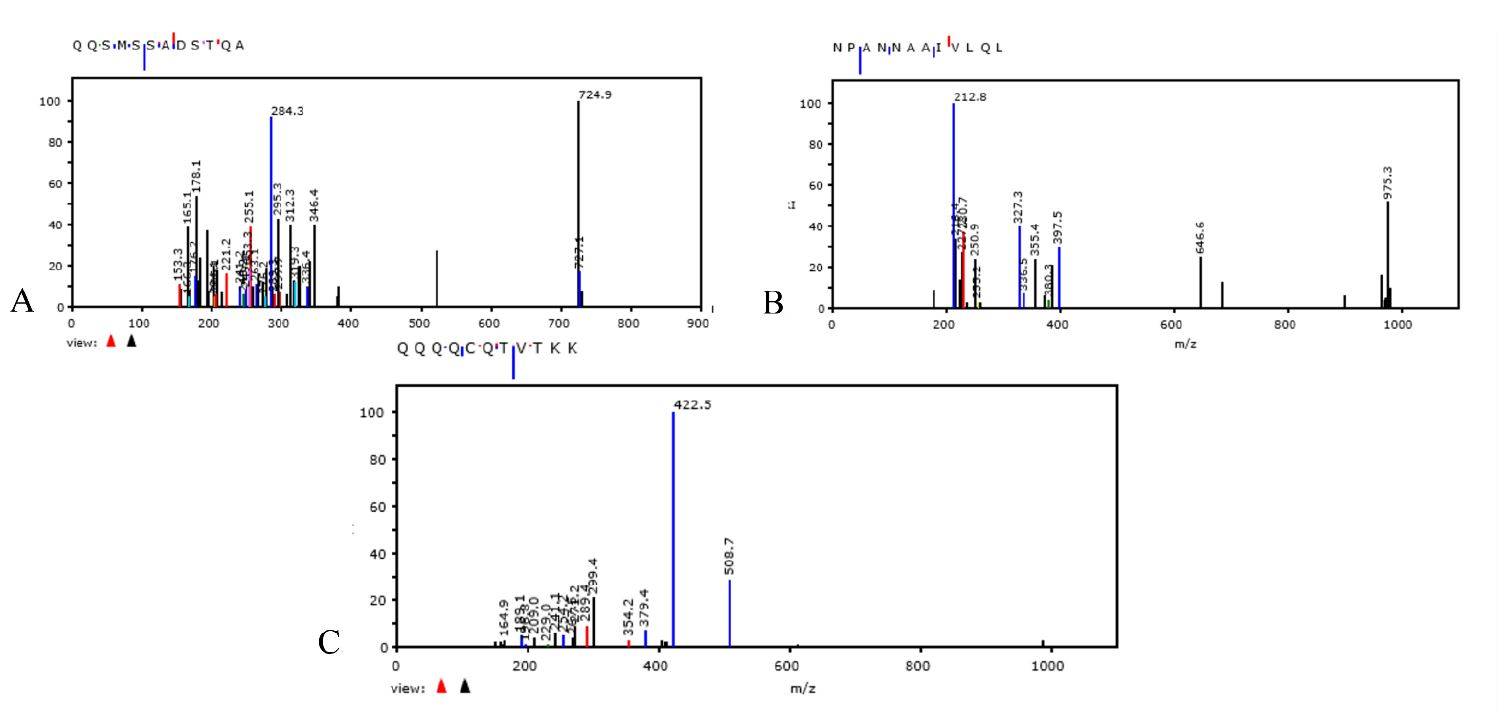
After 30 days of bacterial growth in liquid medium, nitrogen (15N) isotope-labeled medium (Sigma Aldrich) was added. After an additional 7 days, proteomic analysis of SARS-CoV-2 was performed. Modified peptides of the SARS-CoV-2 nucleocapsid protein were detected by extracting the 13C n15N species as specified in Materials and methods The spectra obtained by Tandem mass spectrometry (MS/MS) [16] and acquired between 200 and 1800 m/z (mass/charge), show fragmentation of modified peptide of Nucleocapsid SARS-CoV-2 protein, containing 13C and 15N isotopes. The sequences are shown in figure 4 A-C. The nitrogen isotope (15N) was detected in nucleocapsid peptide of SARS-CoV-2 replicated in bacterial cultures by monitoring the molecular mass shift. The MS/MS spectra shows that the 15N species is located on aminoacid residue of peptides. Nitrogen (15N) isotope labeling in SARS-CoV-2 proteins demonstrates that bacteria can replicate, transcribe, and translate viral RNA and thus that bacteria produce the proteins of this virus.
Figure 3. Panel A: represents the phylum of bacteria present in the culture before the addition of SARSCoV-2 while panel B represents the change in bacterial population after 30 days of the same cultures infected with SARS-CoV-2 .
For more detail see supplementary materials.
Figure 4. Panels A-C: shows the MS/MS spectra of the nucleocapsid protein of SARS-CoV-2 containing
the nitrogen isotope. Panel A shows peptide seq: QQSMSSADSTQA;
ID:|A0A8B1JYE4|A0A8B1JYE4_SARS2; Mods: Q408 + 1 (13C) +2 (15N), Q409 + 1 (13C) +2 (15N) . Panel
B shows peptide seq: NPANNAAIVLQL ; ID: A0A7M1YDW6|A0A7M1YDW6_SARS2;; Mods: Q160+
1 (13C) +2 (15N). Panel C shows peptide seq: QQQQCQTVTKK; ID:|A0A8B1XSI6|A0A8XSI6_SARS2;
Mods: Q239+ 1 (13C) +2 (15N), Q239+Ammonia-loss, Q240+ 1 (13C) +2 (15N), Q241+ 1 (13C) +2 (15N),
Q242+ 1 (13C) +2 (15N), Q244+ 1 (13C) +2 (15N).
3. Discussion
In an effort to counter the current global pandemic, characterization of the molecularlevel
interactions with the host of the novel SARS-CoV-2 coronavirus that causes COVID-19
has become the subject of much scientific attention. However, when the virus enters the body
it also interacts with microorganisms that already inhabit the host. Understanding virus-hostmicrobiome
interactions may provide additional insight into the biological processes
perturbed by viral invasion. Sifting through old scientific literature can be challenging and
valuable. For example, some past studies have observed a co-participation of bacteria in the
infection of another RNA virus such as HIV [17]. In addition, recent studies have observed
the presence of HIV in intestinal mucosal lymphoid tissues[18].
In a recent case report, it is reported that 500 days after recovering from Ebola virus
(fecal-oral RNA virus), an African boy sexually transmitted the virus to a woman [19]. One
would wonder: how is this possible? Where was the Ebola virus hiding?
It should be kept in mind that many studies have observed that SARS-CoV-2 is almost
always present in the feces of sick and asymptomatic individuals, suggesting a fecal-oral route
of transmission[20]. In addition, its closest relatives (coronavirus RATG13) have been found
in the feces of bats [5]. Experience with the SARS-CoV-2 also suggests this route of
transmission [21]. During outbreaks of other coronaviruses, CoV-OC-43 and CoV-NL-63,
district-like epidemics of these viruses have been reported, with the same symptoms and
epidemic statistics as SARS-CoV-2 [22]. The coronaviruses of pigs and cattle also show a
circumscribed transmission pattern [23]. If we look more closely at the history of poliovirus
(yet another RNA virus), it was only after many years that it became clear that its transmission
was not exclusively airborne, as was mistakenly believed, but also fecal-oral [24]. In fact,
observing the epidemiology and spread of polioviruses, it is evident that they have always
caused district epidemics but have never generated a pandemic crisis. It is also well known
that a problem faced by the scientific community was the procurement of sterile artificial steel
lungs for hospitals to contain their spread [25].
In 1956, Albert B. Sabin indicated the pathogenesis of poliomyelitis, which had been
debated until then “ Polio is an RNA virus with fecal-oral transmission. Its significant
replication occurs in the intestinal tract” [26-29].
Interestingly, at that time, a group of researchers observed how bacteria might play a role
in the selection of a few different strains of poliovirus. They observed that bacterial endotoxins
induced a lower rate or even inhibited viral replication [30]. Similarly to our studies [6,7], we
highlighted several toxin-like proteins in the plasma, feces, and urine of COVID-19 patients,
which were produced by bacteria as a consequence of interaction with SARS-CoV-2. We have
shown that these oligopeptides are derived from bacterial cultures [6, 7] and are testing them
in mice (data in preparation). We have already observed the replication of SARS-CoV-2 virus
in bacteria many times [7] and have done some experiments with Nitrogen Isotope 15N in the
bacteria culture in the presence of this virus to better understand this phenomenon (Figure 4).
Furthermore, we found that after 30 days of bacterial cultures, using previously published
methodology [7], some bacterial genera tend to increase and others to decrease. These data
suggest that the increase in viral load, as previously shown by Petrillo et al [7], in bacterial
cultures, is associated with a decrease in some bacterial genera after 30 days, perhaps is due
to the lytic cycle of these bacteria (Table 1 in supplementary material). We found that some
bacterial genera, such as Dorea, Fusicatenibacter, Klebsiella, Streptococcus decreased, while
other genera, such as Campylobacter, Prevotella, Staphylococcus, Bacteroides, and
Cytobacter increased. Our results are in line with those of Liu et al., 2021 [31] that performed
a metabolomic analysis of blood, urine, and nasopharyngeal swabs of a COVID-19 and non-
COVID-19 patient group and a metagenomic analysis of pharyngeal samples. Their results
indicated that nasopharyngeal commensal bacteria including Gemella morbillorum, Gemella
haemolysans, and Leptotrichia hofstadii were significantly depleted in the pharynges of
COVID-19 patients, whereas bacteria such as Prevotella histicola, were relatively increased.
In addition, the mechanism of interaction that we have observed could be very similar to that
previously reported by Ebrahimi, 2020 [11] in which SARS-CoV-2 spike glycoproteinbinding
proteins, e.g. ACE2-like protein, are expressed by commensal bacteria of URT.
However, it must be considered that the interaction between bacteria and SARS-CoV-2
does not necessarily occur with native viral surface proteins, but most likely with a surface
reworked by proteases and toxins produced by bacteria. A likely explanation for this novel
interaction between bacteria and coronaviruses could be based on the highly dynamic process
generated between bacterial products (toxins, metabolytes, oligopeptides, proteases) and viral
proteins.“ In addition, we have observed lytic plaques using bacterial species of the genus
Dorea (see Figure 2 of Supplementary Materials) and have made the method, we have
described, accessible to many researchers for further investigation. Our method demonstrates
that with a fluorescence microscope it is possible to observe in cultured bacterial cells an
increase in viral replication of bacteriophages in general and SARS-CoV-2 in particular (see
figure 3 of the supplementary materials). Therefore, immuno-EM, combined with molecular
analysis of viral growth [7], and complemented by a fundamental assay of bacterial culture
medium in the presence of nitrogen (15N), are important steps to observe and understand the
mechanisms of bacteriophages.
It must be also remembered that bacteria are found on oral, nasal, pharyngeal, alveolar,
and intestinal mucous membranes and are often more numerous than epithelial cells. Even if
a virus does not replicate in them, this does not imply that it will not cause discomfort through
the bacterial flora of the human being. Understanding that a virus also binds to, interacts with,
and infects bacteria and that fecal-oral transmission is an additional source of spread,
completely changes the epidemiological scenario of SARS-CoV-2. It changes how to
intervene, how to give policy input, how to treat COVID-19 patients at the onset of symptoms,
how to prevent, and perhaps another possible solution could be the vaccine as Sabin did: “an
attenuated virus for oral administration”, probably justified by the need of the bacteria of the
microbiota to interact with the virus.
Understanding whether bacteria can engineer certain viruses, can contribute to new
mutations, as we [6, 7], and others have reported [26], can have their own defense
mechanisms, can react by producing toxic molecules to defend themselves [6], whether that
defense ceases or remains in perpetuity, require a number of essential controls to be
performed. These controls should be conducted more logically if the pathogen causes so much
social and community damage.
Any treatment or use of preventive medical approaches without these controls can lead to
failures, especially if the correct diagnosis is not made.
The pandemic began in December 2019, and the scientific community still does not agree
on when the virus began spreading among humans and what is the best solution. It is crucial
to consider that once we have demonstrated the replication of this virus in bacteria and
understood that the route of transmission is also fecal-oral, could be possible to control the
virus spread and contribute with also other specific pharmacological therapies to support
current vaccines in order to improve the vaccination campaign.
4. Materials and Methods
In vitro bacterial cultures were obtained from fecal (stool) samples from subjects both
SARS-CoV-2 positive and negative (for control cases) for viral infection. Informed consents
from patients were obtained according to local legislation. Replication of virus in human
bacteria by statistical analysis, Luminex molecular tests, and further controls by mass
spectrometry technique (SACI technique) are reported in Petrillo et al., 2021 [7]. Immunogold
was performed with the bacteria culture pellet at day 30 after centrifugation of the aliquots.
Supernatants were used from the same samples to acquire immunofluorescent images using a
fluorescence microscope (100X magnification, Zeiss Axioplan 2, Axiocam 305 color
supplementary materials). Each step was repeated three times.
4.1. Electron microscopy
For electron microscopy analyses, aliquots of bacterial pellets (0.5 mL) obtained after 30
days of bacterial cultures were fixed by resuspending them in 1% glutaraldehyde. They are
then centrifuged at 10000 rpm for ten minutes, harvested as pellets, fixed in 2% OsO4, and
processed according to the standard procedures, by dehydration in ethanol followed by
infiltration and embedding in Epon resin and polymerization at 60°C for 48 hours. 60-nm thin
sections were cut from the sample using a Reichert-Jung ultramicrotome. Grids were
counterstained with UranyLess EM stain and Lead Citrate 3% (Electron Microscopy Science).
Images were obtained from thin sections under an electron microscope (Tecnai G2 Spirit
BioTwin; FEI) equipped with a VELETTA CCD digital camera (Soft Imaging Systems
GmbH).
4.2. Immunogold labelling tecnique
Another pellet of 0,5 mL of the same bacterial cultures was fixed by resuspending with
0.05% glutaraldehyde and 4% paraformaldehyde in 0.1 M PBS buffer, and then centrifugate
to 10000 rpm for ten minutes. Thereafter, the bacteria pellets were post-fixed in 1% OsO4, and
processed according to the standard procedures, by dehydration in ethanol followed by
infiltration and embedding in Epon resin and polymerization at 60°C for 48 hours. Ultrathin
(60-nm) sections were cut from the sample using a Reichert-Jung ultramicrotome and
collected onto Formvar/Carbon coated nickel grids. After etching with 3% sodium meta
periodate and blocking with 3% BSA in PBS buffer, the samples were incubated in a primary
rabbit monoclonal [EPR24334-118] to SARS-CoV-2 nucleocapsid protein antibody (Abcam
ab 271180) 1:1000 in 1% BSA in PBS buffer overnight at 4°C and then in anti-rabbit 10 nm
gold-conjugated secondary antibody (Aurion) 1:20 in PBS buffer for 2 hours at room
temperature.
Negative control was obtained by omitting the primary antibody and using only the
secondary gold-conjugate antibody. The control of immunogenicity of antibodies primary
versus nucleocapsid protein of SARS-CoV-2 was assumed from Shang C. et al. [32].
Grids were counterstained with UranyLess EM stain and Lead Citrate 3% (Electron
Microscopy Science) and images were acquired under a transmission electron microscope
(FEI Tecnai G2 S-TWIN) equipped with a bottom-mounted FEI Eagle4k digital camera.
4.3. Immunogold, pre-embedding technique.
Bacteria pellets (0,5 mL) were fixed by resuspending with a mixture of 0.05% glutaraldehyde
of 4% paraformaldehyde in 0.2M HEPES buffer. Then bacteria pellet, obtained by
centriguation ( 10000 rpm for ten minutes), were resuspended, washed with PBS, and
incubated first in blocking solution (50mM NH4Cl, 0.1% saponin, 1%BSA in PBS) and then
in primary antibody against Nucleocapsid SARS-CoV-2 (N) protein (Abcam, #ab273167).
Then the cells were washed with PBS and incubated with NANOGOLD conjugated anti-rabbit
Fab fragments (Nanoprobes, #2004). After washes in PBS and distilled water bacterial pellets
were kept in the gold-enhancement mixture (Nanoprobes). Immunolabelled specimens were
post-fixed in OsO4 and uranyl acetate and embedded in EPON. Thin sections were cut from
embedded specimens using Leica EM UC7 (Leica Mycrosystems). Electron microscopy
images were acquired from thin sections under an electron microscope (Tecnai G2 Spirit
BioTwin; FEI) equipped with a VELETTA CCD digital camera (Soft Imaging Systems
GmbH).
4.3.1 Validation of antibody against Nucleocaspid-protein of SARS-CoV-2 with fluorescent microscopy
cDNA encoding green fluorescent protein (GFP)-tagged N-protein of SARS-CoV-2 was
generously provided by Dr. M.A. De Matteis (TIGEM, Pozzuoli, Italy) and transfected into
HeLa cells using TransIT-LT1 reagent (Mirus Bio LLC) according to the manufacture
instruction. Transfected cells were fixed in 4% paraformaldehyde, washed with PBS and
incubated in blocking solution (50mM NH4Cl, 0.1% saponin, 1%BSA in PBS). Then the cells
were labelled using primary antibody against N protein (Abcam, #ab273167) and secondary
anti-rabbit IgG conjugated with Alexa Fluor 568 (Thermo Fisher; # A-11011). After labelling
the samples were examined under a LSM700 confocal microscope (Zeiss). Efficiency of
antibody against N-protein was confirmed based on the presence of Alexa Fluor 568 signal in
the cells expressing GFP-tagged N-protein.
4.4. Immunofluorescence
Aliquots of the supernatants of the samples were obtained and the slices were fixed with
4% PFA for 5 minutes. After permeabilization with Triton 0.3% in PBS for 10 minutes, the
slices were rinsed in PBS and blocked with BSA 1% and saponin 0.05% for 30 minutes. The
slices were incubated with the primary antibodies for 2 hours at room temperature and then
with secondary antibodies for 1 hour at room temperature. Immunofluorescence were
performed according to the manufacturer’s protocol using a primary antibody against the virus
nucleocapsid protein („Sars Nucleocapsid Protein Antibody [Rabbit Polyclonal] – 500μg 200-
401-A50 Rockland“), the „Goat anti-RabbitIgG (H+L) Cross-Adsorbed Secondary Antibody,
Cyanine3 #A10520″ as a secondary antibody, and a primary antibody against gram-positive
bacteria („Gram-Positive Bacteria Ab (BDI380), GTX42630 Gene Tex“) and „Goat anti-
Mouse IgG (H+L), Super-clonal™ Recombinant Secondary Antibody, AlexaFluor 488″ as a
secondary antibody“. The negative control of the bacterial cultures was performed without
primary antibodies versus gram-positive bacteria and without primary antibodies versus
nucleocapsid SARS-CoV-2 proteins. The control of immunogenicity of primary antibodies
versus bacteria Gram-positive was performed on the culture negative at Luminex molecular
test to SARS-CoV-2. It was also assume the control of immunogenicity of antibodies versus
Gram-positive bacteria from Kohda et al. and Kameli et al. [33-34]. The control of
immunogenicity of primary antibodies versus nucleocapsid protein of SARS-CoV-2 was
assume from Zhao et al. [35].
4.5 Proteomic profiling analysis
One bacterial liquid culture was used to add, at the end of 30 days, 0,20 gr. of a nitrogen
isotope (15N) labeled medium ( Sigma Aldrich). After another 7 days, a SARS-CoV-2
proteomic analysis was performed, on aliquots (0,2 mL) of the same samples by SACI
technology [16]. Modified peptides of nucleocapsid SARS-CoV-2 protein were detected by
extracting the species 13C n15N ( where n>1). Ion proton rearrangement reaction occurring in
Collisional Induced Dissociation (CID) conditions were considered in the database search data
elaboration [36].The ion were isolated using 5 m/z units and the fragmentation energy was
35% of its maximum value (5V peak to peak).
4.6 NGS sequencing data
DNA from fecal samples, at day 0 (B0) was extracted with E.Z.N.A.® Stool DNA Kit (Omega
Bio-Tech, GA, USA). Two types of kit for the nucleic acid extractions from samples after
30 days (B1) of culture were used: MasterPure Complete DNA and RNA purification kit
(Lucigen, WI, USA) and PureLink Viral RNA/DNA kit (Thermo Fisher Scientific, MA,
USA). All extractions were performed following the manufacturers‘ recommendations. The
Ovation® Ultralow V2 DNA-Seq Library Preparation kit (NUGEN, San Carlos, CA) was
used for library preparation, following the manufacturer’s instructions. Both input and final
libraries were quantified with the Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA), and
quality was tested by Agilent 2100 Bioanalyzer High Sensitivity DNA assay (Agilent
Technologies, Santa Clara, CA). Libraries were then prepared for sequencing and sequenced
on NovaSeq 6000 in 150-bp paired-end mode. Bioinformatic reconstruction was calculated
on the fasta file of bacterial sequences on UniProt. Result are presented in Table 1, in
supplemetary material. In addition to exlude other bacteriophage viruses particle the
bioinformatic reconstruction was calculated also on the fasta file of bacteriophage sequences
on UniProt (Table 2, in suplementary materials).
5. Conclusions
The control of prokaryotic cells as a reservoir source for some viruses represents an
element that should be the basis of any diagnostic pathway regardless of whether one is
convinced that it can occur or not. Our results obtained by Immuno-EM, embedded
immunogold, and fluorescence imaging highlight a close connection between the coronavirus
SARS-CoV-2 and bacteria of the intestinal microflora. SARS-CoV-2 has two mechanisms of
action; it infects both the eukaryotic cell, as reported in the current literature, but it also infects
the human bacterial flora (two bacteria are already identified, data in preparation) and Sabin’s
solution, the attenuated vaccine for oral administration, could be a solution that could
complement the vaccination campaign to exit definitively from this pandemic. Our results
confirm the role of bacterial co-factors in the multiplication of COVID 19 coronavirus during
this new epidemic confirmed by the efficacy of some previously described antibiotics [7,8]
on the early phase of viral infection. Whether it also plays a role in long-term disease remains
to be determined. In this case, we could consider the virus as a bactériophage with a lytic
phase and a lysogenic phase. Furthermore, the presence of the nitrogen isotope 15N in viral
proteins after infection of bacteria with SARS-CoV-2 could represent a new approach to study
the interaction of RNA viruses with human microflora.
Author Contributions: “Conceptualization, C.B. and M.P. (Marina Piscopo); methodology, C.B. D.B. L.F.; software, C.B, B.B., L.F.; validation, M.P., S.C.; formal analysis, S.C. and M.P.( Marina Prisco); investigation, C.B:, S.C..; resources, C.B. and G.M.; data curation, M.P.( Marina Piscopo) and C.B..; writing—original draft preparation, C.B. and M.P..; writing—review and editing, M.P..; visualization. M.P., and C.B..; supervision, C.B., and M.P.; project administration, C.B..; funding acquisition, G.M., C.B. All authors have read and agreed to the published version of the manuscript.”
Funding: Please add: “This research received no external funding”
Institutional Review Board Statement: the study did not require ethical approval.
Informed Consent Statement: “Informed consent was obtained from all subjects involved in the study.”
Data Availability Statement: “Not applicable” here.
Acknowledgments: I thank Dr. M. Petrillo, Dr. G. Ciammetti, Prof. O. Piazza (University of Medicine in Salerno). I also thank the Biogem Institute of Ariano Irpino (Av) for fluorescence microscope images. We would like to acknowledge TIGEM Advanced Microscopy and Imaging Core for the help with preparation of specimens for fluorescent and electron microscopy. We also are grateful to the Electron microscopy facility of the department of chemical sciences- University of Study Federico II Naples (UniNa). I also am grateful to Marsan consulting and Dr. Marino Giuliano for their full support.
Conflicts of Interest: “The authors declare no conflict of interest.”